New Service: ATAC-sequencing
ATAC-seq (Assay for Transposase-Accessible Chromatin sequencing) is a technique used to assess genome-wide chromatin accessibility now available at Genomics & RNA Profiling Core (GARP). This cutting-edge technology empowers researchers to explore genome-wide chromatin accessibility, providing unprecedented insights into gene regulation, transcription factor binding, and epigenomic landscapes.
To find out more about this service, contact us by email at garpcore@bcm.edu.
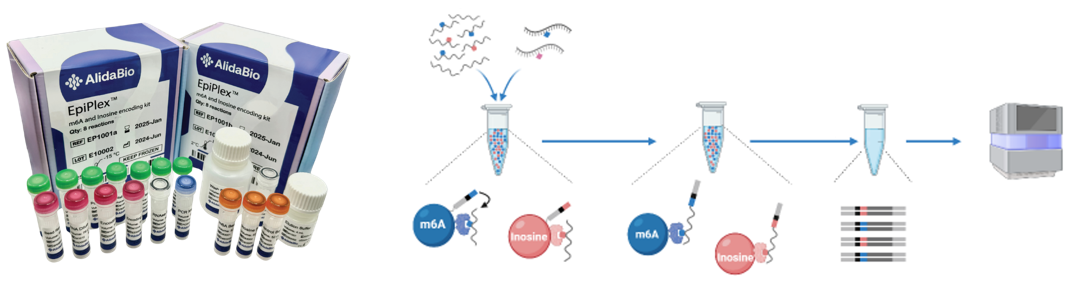
New Service: Multiplexed RNA modification sequencing
The GARP core partnered with the AlidaBio to offer a new service which is multiplexed detection of RNA modification. The assay identify variations in both the total abundance and the specific locations of m6A and inosine modifications.
To find out more about this service, contact us by email at garpcore@bcm.edu.
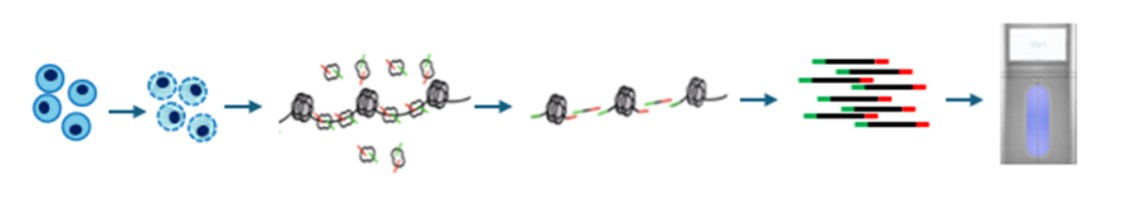
New Service: Genome-wide histone PTM profiling by CUT&RUN- Sequencing
We now provide end-to-end CUT&RUN (Cleavage Under Target and Release Using Nuclease) service, from cells through data.
To find out more about this service, contact us at garpcore@bcm.edu.
About the Core
The Genomic and RNA Profiling Core (GARP) provides access to state-of-the-art genomic profiling technology and services, including:
- Sequencing only- Standalone sequencing of user-prepped samples as opposed to samples that go through library prep and sequencing in-house.
- Library prep
- Next Gen Sequencing on our Illumina platforms (iSeq 100 and NovaSeq X)
- Long Read sequencing on the Oxford Nanopore PromethION 24 platform
- NanoString nCounter Technology
- Sample Quality Control
- Nucleic acid shearing on the Covaris
- DNA size selection on the Pippin HT
- Consultation
- Primary Data Processing & Analysis
Core Resource
We serve as the core resource for the following:
Information for Acknowledgement of Cores in Publications
Core name: Genomic & RNA Profiling Core
Personnel: Daniel C. Kraushaar, Ph.D., Director; Kieu Pham, Ph.D., Sr. Staff Scientist; Emily Ricco, Lab Manager; Stacy Nguyen; Ping Kang; Esmanur Tokcan; Angelinda Maria Maldonado, Research Staff.
Grants supporting the core: P30 Digestive Disease Center Support Grant (NIH-NIDDK - 5P30DK056338) and P30 Cancer Center Support Grant NIH NCI (P30CA125123) and CPRIT (RP200504), P30 Center for Precision Environmental Health (NIEHS-5P30ES030285-03), NIH S10 grant (1S10OD036427).
The associated centers should adhere to these additional acknowledgement instructions.