What is Chromatin? What are Histones?
The physiological state of eukaryotic genomes is the chromosome. Chromosomes are approximately 50% protein by weight. This nucleic acid-protein complex is referred to a chromatin. Histones are highly basic proteins that make up the largest bulk of the protein component of chromatin. There are four classes of core histones: H2A, H2B, H3 and H4. In addition there is a linker histone: H1. Within these classes there are often multiple members or variants of the class. Histones form the core of the nucleosome which are the fundamental repeating unit of chromatin. The protein component of chromosomes plays an essential role in genome biology, often regulating expression of gene. Posttranslational modifications (PTMs) of histones have many known functions in the biology of the genome; however, there is much yet to be understood. We are particularly interested in how chromatin appears to function in complex multivalent ways. That is more than one source of variability is operating in tandem to transduce a signal or recruit an activity. For example, most of the ‘readers’ of chromatin (proteins with domains that site specifically recognize PTMs) usually have two domains that read more than one PTM simultaneously. In this way signals may be integrated and more complex information stored and retrieved.
Histone Proteoforms
Histones are densely posttranslationally modified. That is that the normal state of histones is to be multiply modified rather than being mostly unmodified with an occasional PTM occurring in low abundance. This results in a plethora of combinations (or proteoforms) defining many different species. With tens of PTMs on a single protein molecule there can be trillions of possible ways in which they can combine to define unique species (proteoforms). Because chromatin functions in a multivalent manner and because histone PTMs are central to genome regulation and genome dysregulation is central to many disease, especially cancer, we are interested in accurately and sensitively measuring histone proteoform in an unbiased, quantitative manner.
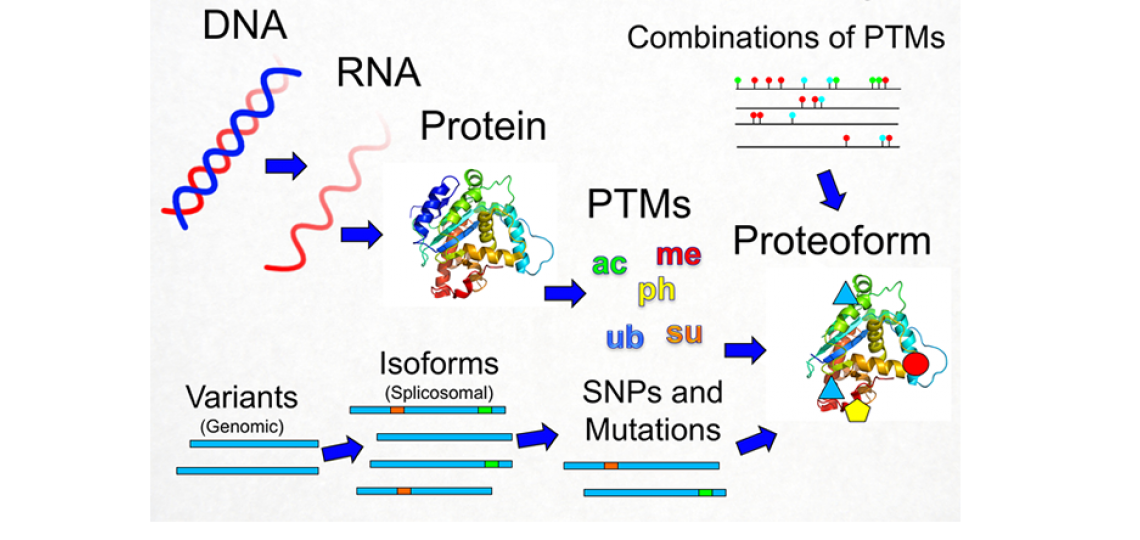
The Proteoform: The Next Level of Protein Biochemistry
Figure 1: Illustration of the Proteoform: A proteoform is a single molecule protein species where all sources of variation (PTMs, SNPs, etc.) are unambiguously defined in a specific combination.
Top Down Proteomics
Top down proteomics forms the core technology focus of our laboratory. Although technologically connected and operating within similar biological scopes, top down proteomic fundamentally answers different questions than its more familiar cousin ‘bottom up’ proteomics. Most importantly to us is that top down proteomics is capable of measuring proteoforms. Because we introduce the sample intact into the mass spectrometer the connectivity of PTMs (or other sources of variation) is preserved, even if they are on distal ends of the protein. We use the cutting edge in mass spectrometry technology and innovative chromatography to physically separate, gas phase isolate, fragment and ultimately sequence and identify proteoforms.








