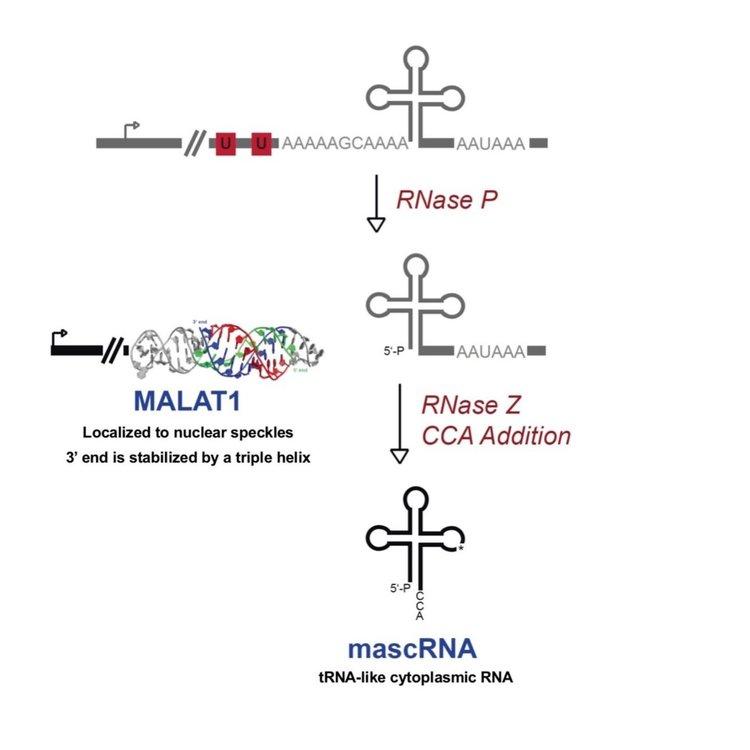
Much of our early work focused on the MALAT1 locus, which is over-expressed in many human cancers and produces a long nuclear-retained noncoding RNA as well as a tRNA-like small RNA known as mascRNA, MALAT1-associated small cytoplasmic RNA. Despite being an RNA polymerase II transcript, the 3’ end of MALAT1 is produced not by canonical cleavage/polyadenylation but instead by recognition and cleavage of the tRNA-like structure by RNase P (Figure 4).
The MEN β long nuclear-retained noncoding RNA, also known as NEAT1_2, is similarly processed at its 3’ end by RNase P. Surprisingly, although processing of MEN β produces a tRNA-like small RNA, it is generally rapidly degraded in vivo. Pursuing this observation, we revealed a universally conserved quality control mechanism mediated by the CCA-adding enzyme. Normally, the CCA-adding enzyme adds CCA to the 3’ ends of tRNAs, a critical step in tRNA biogenesis that generates the amino acid attachment site. However, the enzyme adds CCACCA to structurally unstable tRNAs and tRNA-like small RNAs (including the MEN β tRNA-like transcript and certain hypomodified tRNAs) marking them for degradation. We conjecture that CCACCA addition prevents errors in translation and controls tRNA levels, an especially critical function considering that even slight changes in tRNA levels can drive cell proliferation and oncogenic transformation.
As the mature 3’ ends of MALAT1 and MEN β are generated by RNase P and not by the cleavage and polyadenylation machinery, these long noncoding RNAs lack poly(A) tails. Nevertheless, these RNAs are expressed at levels higher than many protein-coding genes. We showed that the 3’ ends of these noncoding RNAs are protected from 3’-5’ exonucleases by highly conserved triple helical structures. Surprisingly, when these triple helical structures are placed downstream from an open reading frame, the transcript is efficiently translated in vivo despite the lack of a poly(A) tail. This result challenges the common paradigm that long poly(A) tails are required for RNA stability and efficient protein synthesis and suggests that non-polyadenylated RNAs may produce functional peptides in vivo via mechanisms that are likely independent of poly(A) binding protein.
Key Publications
-
Doucet, A.J., Wilusz, J.E., Miyoshi, T., Liu, Y., and Moran, J.V. (2015) A 3’ poly(A) tract is required for LINE-1 retrotransposition. Mol Cell 60: 728-741
-
Kuhn, C.-D., Wilusz, J.E., Zheng, Y., Beal, P.A., and Joshua-Tor, L. (2015) On-enzyme refolding permits small RNA and tRNA surveillance by the CCA-adding enzyme. Cell 160: 644-658.
-
Wilusz, J.E., JnBaptiste, C.K., Lu, L.Y., Kuhn, C.-D., Joshua-Tor, L., and Sharp, P.A. (2012) A triple helix stabilizes the 3’ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev 26: 2392-2407.
-
Wilusz, J.E., Whipple, J.M., Phizicky, E.M., and Sharp, P.A. (2011) tRNAs marked with CCACCA are targeted for degradation. Science 334: 817-821.
-
Wilusz, J.E. and Spector, D.L. (2010) An unexpected ending: non-canonical 3’ end processing mechanisms. RNA 16: 259-266.
-
Sunwoo, H., Dinger, M.E., Wilusz, J.E., Amaral, P.P., Mattick, J.S., and Spector, D.L. (2009) MEN ε/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19: 347-359.
-
Wilusz, J.E., Freier, S.M., and Spector, D.L. (2008) 3’ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135: 919-932.








