Altered hedgehog signaling is now implicated in the development of approximately 20-25 percent of all cancers, especially soft tissue cancers. In most cases, it appears that hedgehog activation plays a causal role. Network activation can be achieved by a number of gain-of-function mutations in positively acting components (e.g. ligands, Smo, Gli1, Gli2), as well as by loss-of- function mutation in Ptch1, a key inhibitor of hedgehog network function, as well as in Su(Fu).
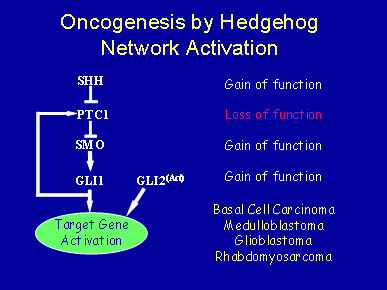
Several genes in the mammalian hedgehog signaling network are known oncogenes or tumor suppressor genes. For example, loss-of-function mutations in Ptc1, or gain-of-function mutations in Smo, Shh, Gli1, and Gli2, can each contribute to skin cancers, most notably basal cell carcinomas – the most common of all human cancers. In addition, Ptc1 has been implicated in the development of medulloblastomas, glioblastomas, rhabdomyosarcomas, pancreatic cancers, and other soft tissue tumors. Recently, small cell lung cancer was shown to be dependent on activated hedgehog signaling. In some cases, mutations in hedgehog network genes have not been identified, leading to the possibility that hedgehog network genes need not be mutated to contribute significantly to cancer progression. Rather, the network may need only be inappropriately regulated.
Mutational Analysis
In mutational analysis, an early study found Ptch1 mutations in 2 of 7 human breast cancers (Xie et al. 1997). Additionally, a Ptch1 polymorphism was linked to increased breast cancer risk associated with oral contraceptive use (Chang-Claude et al. 2003). More recently, array comparative genomic hybridization (CGH) analyses indicate that genomic loss at the Ptch1 locus was the fourth most commonly detected change among the tumor suppressor genes identified in the study, occurring in 19 percent of human breast cancers and 33 percent of breast cancer cell lines (Naylor et al. 2005). However, until very recently (Sjoblom et al. 2006), no mutations in other network components had been identified in breast cancer (Vorechovsky et al. 1999). With exhaustive sequencing efforts, 3 missense mutations in the Gli1 gene were identified in 11 breast cancer samples examined, but these mutations were carried in the heterozygous state, and have not yet been analyzed functionally.
By expression analysis, a recent immunohistochemical staining study suggested that hedgehog signaling is activated in a majority of human invasive breast cancers based on ectopic expression of PTCH1 and GLI1 (Kubo et al. 2004) in 50 out of 52 clinical samples investigated. However, as mentioned previously, neither SHH, PTCH1 nor GLI1 expression were detected in normal tissue in this same study, whereas these genes have been detected in normal tissue by others. For example, a second study examining differential expression of hedgehog network genes in at least 8 patient matched samples total showed SHH, PTCH1 and GLI1 expression (Mukherjee et al. 2006) in both normal and cancer tissue. In this study, expression of SHH was increased >2-fold in the epithelium of 5 of 8 samples, with a >2-fold increase in GLI1 expression in cancerous epithelium in 4 of 8 samples examined. There was also a >2-fold increase in PTCH1 expression in 3 of 9 samples examined. Immunostaining of these three proteins in normal or cancer-associated stroma was reduced relative to the epithelium. Using microdissection, Gli1 protein and mRNA expression were positively correlated with one another. However, Ptch1 protein and mRNA expression were not positively correlated in these samples, as might be expected based on current models for hedgehog network function. With respect to mRNA expression, one striking observation was that Smo mRNA levels were significantly higher than normal in 4 of 10 samples examined.
We have also conducted an immunohistochemical study for expression of PTCH1 and SMO in a large panel of normal, ductal carcinomas in situ (DCIS), and invasive breast cancer (IBC) samples (Moraes et al., 2007). In our study, like that of Mukherjee et al. (Mukherjee et al. 2006), PTCH1 was detectable at moderate levels throughout the epithelium, and in isolated stromal cells of the normal breast. While some samples in our study showed strong PTCH1 expression, PTCH1 expression was decreased or absent in ~50 percent of ductal carcinoma in situ (DCIS) and invasive breast cancers (IBC). With respect to SMO, expression was undetectable in normal breast, but SMO was ectopically expressed in ~70 percent of DCIS and ~30 percent of IBC. PTCH1 expression was not correlated significantly with SMO expression in either DCIS or IBC. Altered SMO protein expression in human breast cancer was generally consistent with the Q-PCR results of Mukherjee et al. (Mukherjee et al. 2006) that showed increased mRNA expression in ~40 percent of their samples. Together, these data suggest altered hedgehog signaling occurs early, and often, in human breast disease progression.
With respect to a possible predictive or prognostic utility of altered hedgehog network gene expression, expression of PTCH1 or SMO did not correlate with histological grade (DCIS only) or with expression of any clinically-relevant marker tested (estrogen receptor, ErbB2, p53). It will be of considerable interest to determine whether SMO or PTCH1 expression correlates with any other clinically-relevant parameter such as metastatic or osteolytic behavior (Sterling et al. 2006) given the recent finding that expression of Gli2 in MDA-MB-231 cells mediates osteolytic behavior of these cells via induction of parathyroid hormone-related protein.
Other than these studies, the strongest evidence for a role in breast cancer comes from the published genetic studies in mice showing hyperplastic defects in mammary glands of ΔPtc1/+and ΔGli2/ΔGli2 mutants (Lewis et al., 1999; Lewis et al., 2001), as well as in transgenic mice expressing activated SMO under the control of the MMTV promoter (Moraes et al, 2007).








