
High Throughput Proteoform Analysis
We specialize in high throughput top down liquid chromatography-mass spectrometry. This allows us to study chromatin biology with proteoform level specificity and observe quantitative changes to reveal mechanisms of disease.
About the Lab
Our laboratory works at the intersection of three fields: epigenetics, top down proteomics and cancer. We use cutting edge top down proteomics tools to study the proteoforms of histones, transcription factors, nuclear receptors, co-activators/co-repressors, and chromatin ‘readers’, ‘writers’ and ‘erasers’. This proteoform level analysis of chromatin provides new insights into how chromatin regulatory mechanisms work and inform potential therapeutic strategies for cancer.

Chromatin Biology and Transcription Regulation
Chromatin is the physiological state of the genome. Approximately 50% of chromosomes and chromatin is protein. These include histones, transcription factors, nuclear receptors, co-activators, co-repressors, histone modifying enzymes, nucleosome binding proteins, etc. We study all of the protein components of chromatin with an emphasis on understanding how multiple sources of variation function in concert or combination with each other.
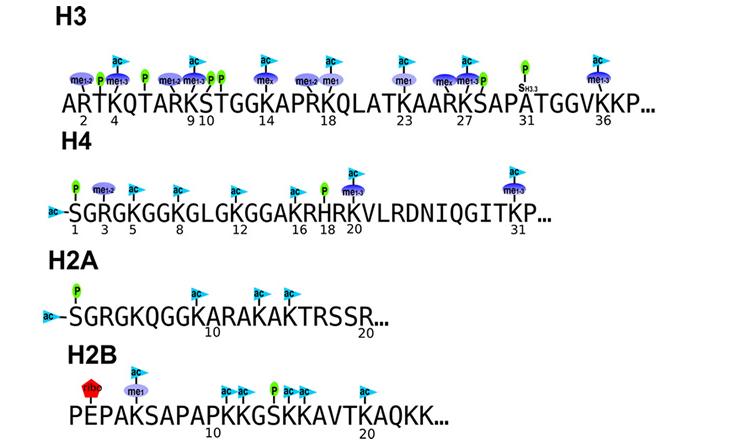
Histones
A major, but not exclusive focus of our lab is the analysis of histones, their variants, post-translational modifications and proteoforms. Histones are essential regulators of the human genome and their dysregulation is central to cancer biology. Yet, due to technical challenges many variants are unstudied and how their dense post-translational modifications function in concerts is still emerging.

Top Down Mass Spectrometry
Top Down Mass Spectrometry enables us to distinguish and quantitate individual proteoforms. Thus, we can definitively identify how combinations of post-translational modifications co-exist on the single molecule level even when they are at distal sites. Furthermore, we can distinguish the specific variants on which these events occur.
LC-MS
By using sophisticated chromatography designed to separate proteoforms that are often structural isomers, we have improved our sensitivity and enabled the observation of otherwise inaccessible variants, post-translational modifications and the dynamics of individual proteoforms.
Lab Projects
- Histone Proteoform Dynamics in Response to Epigenetic Inhibitors
- Histone Variants in Cancer and Cancer Treatment, Transcription Factor Proteoforms,
- Cancer Drug Resistance
- Pharmacological Profiling
- Histone Proteoform Response to DNA Damage
- Computational Approaches to Analyze Top Down Data and Interpret Proteoform Meaning
We are working on a variety of projects that are deeply connected by design and each informs the other in the underlying biological mechanisms.
Dr. Nicolas Young
Visit Dr. Young's Faculty Profile.









