
Submission Data
The center has received 139 submissions for disease modeling, encompassing 191 genetic variants. Half of our submissions are from individuals or programs not affiliated with Baylor College of Medicine.
Currently, 60 submissions, encompassing 75 gene variants, have been accepted into the center, 66 submissions were not accepted, seven submissions are pending final review, and six submissions are on hold.
Data updated: March 10, 2025.
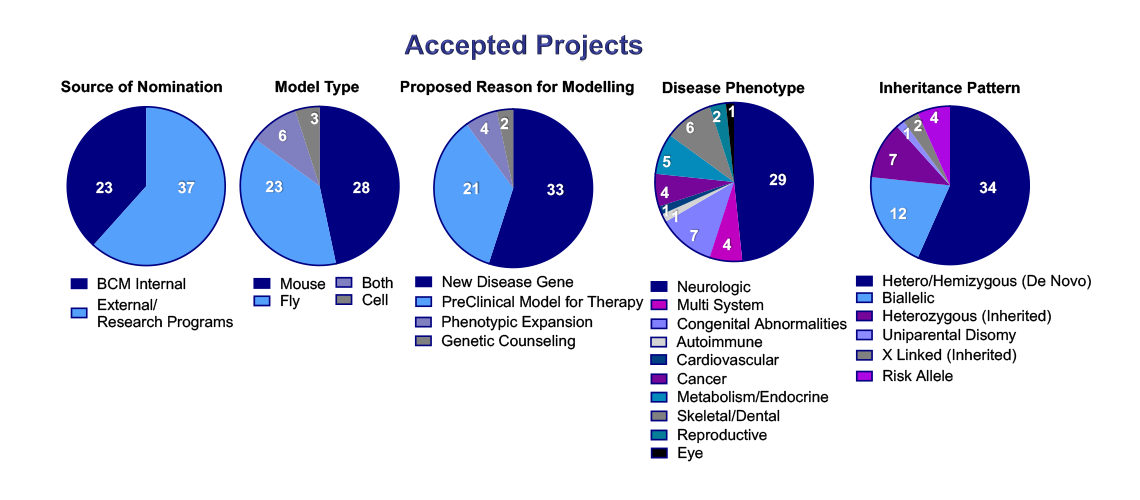
Accepted Projects Data
Source of Nominations
Of the 60 submissions accepted, 37 were from individuals or programs not affiliated with Baylor College of Medicine. The 23 Baylor-affiliated submissions involve collaborations with individuals or programs at other institutes.
Model Types
Of the accepted projects, 28 of the accepted projects use mouse models, 23 use fly models, six use both fly and mouse models, and three use patient-derived cells.
Proposed Reasons for Modeling
Of the accepted projects, 33 projects are using precision modeling to confirm new disease gene discoveries, 31 are employing preclinical precision models for therapy, four are utilizing precision models for validating phenotypic expansion, and two are using precision modeling to assist with genetic counseling.
Disease Phenotyping
These accepted projects involve disease phenotyping for a variety of conditions and organ systems, including neurologic (29), multi-system (4), congenital abnormalities (7), autoimmune (1), cardiovascular (1), cancer (4), metabolism (5), skeletal/dental (6), reproduction (2) and eye (1).
Inheritance Patterns
Of the accepted projects, 34 involve modeling diseases associated with heterozygous (de novo) variants in single genes, 12 involve modeling inherited biallelic variants in single genes, seven involve inherited heterozygous variants in single genes, one involves uniparental disomy, two involve X-linked variants in single genes, and four involve modeling of disease risk alleles.
Data updated: March 10, 2025.
Reasons for Acceptance
The following is a list of reasons why variants were accepted for modeling by the center:
- Generation of a preclinical model for planned therapy studies (e.g., gene therapy, ASO, nutritional intervention)
- Modeling of variants in mouse for phenotypic studies that will provide evidence for a novel disease gene in pediatric or adult populations
- Modeling of variants in fly for studies of the functional impact of variant on protein function to provide evidence of a novel disease gene in pediatric or adult populations
- Modeling of a variant in a known disease gene in the mouse to provide evidence of phenotypic expansion
Reasons for Rejection
The following list contains examples of reasons why variants were not selected for modeling by the center:
- Inability to faithfully model variant due to technical limitations
- Variant of uncertain significance in a known disease gene where phenotype is consistent with known disease phenotype, and there were no other plans for studying the model once phenotyping was completed
- Variant involved a poorly conserved residue and bioinformatic tools did not provide strong evidence of pathogenicity
- Identification of a similar variant in the literature upon review by the Center, and model was no longer deemed necessary by submitter
- Multiple matches in GeneMatcher or other databases such that the model was no longer deemed necessary by submitter
- Identification of a suitable research project with the ability to provide a quicker answer with regards to whether the variant impacts protein function
- Homozygous common variant in a gene where knockout mouse has previously been generated, and mouse phenotype was not similar to human phenotype
- Low percentage of phenotypic mice observed in established mouse models of other variants in the same gene
In many cases, the Center refers variants deemed unsuitable for modeling in the center to other research projects that may be more appropriate for answering the question(s) posed by the submitter.








