



Uncovering the molecular cues that control ribonucleoprotein (RNP) assemblies
Ribonucleases are ubiquitous and drive diverse molecular pathways to promote cellular fitness and survival. RNA processing and decay often requires a series of sequential chemical reactions to convert substrate into product while preventing the accumulation of deleterious intermediates. To ensure accurate and efficient chemistry, RNA editing enzymes often assemble into multi-component molecular machines (i.e. ribonucleoprotein assemblies). While our understanding of the mechanisms coordinating these machines are limited, technological advances, such as the “resolution revolution” in cryo electron microscopy (cryo-EM), has made it possible to uncover their structural organization, function, and regulation with unprecedented detail.
To understand how RNA processing and decay complexes are dynamically regulated, we combine hybrid structural biology (i.e. cryo-EM, X-ray crystallography, SAXS, XL-MS) with enzymology, molecular biology, and cell biology. This multidisciplinary research program uncovers the regulation of ribonucleoprotein assemblies at the atomic, molecular, and cellular level.
Mitochondrial RNA Decay
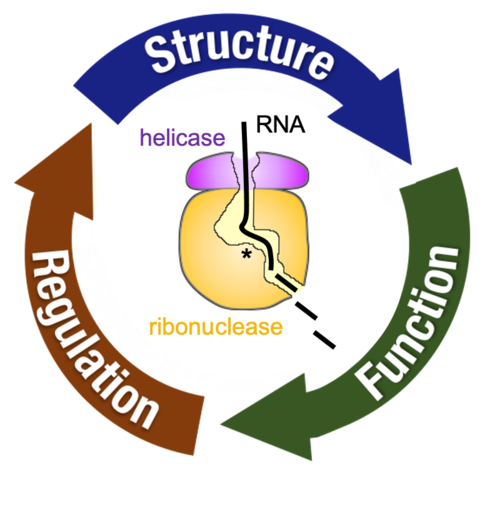
Mitochondria harbor numerous ribonucleoprotein assemblies devoted to RNA processing and decay. The significance of these higher-order complexes is underscored by their link to mitochondrial pathologies including neurological disorders, cancers, and metabolic diseases. While mitochondrial RNA editing factors are coming to light, the mechanism controlling the bulk of mitochondrial RNA (mtRNA) decay remains poorly defined. We apply an integrated research program to investigate the mitochondrial RNA decay machinery, composed of an RNA helicase and ribonuclease. Our work will uncover links between mitochondrial RNA turnover and human disease.








