Consultation
The Multi-Omics Data Analysis Core provides consultation on multiple topics prior to analysis:
- Consultation on experimental design
- Consultation on integration of CPRIT and other core facilities data
- Consultation on integration of publicly available data
- After completion of analysis, review results with primary investigator
Primary Analysis
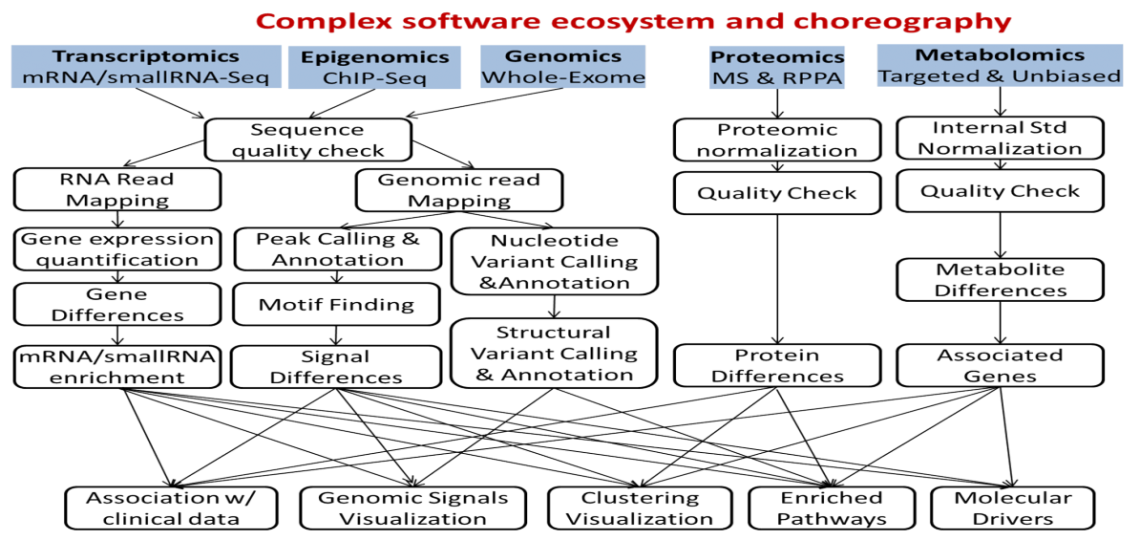
The Multi-Omics Data Analysis group performs primary analysis for the following data types generated in the CPRIT funded cores:
- MS Metabolomics and Lipidomics
- MS Proteomics
- RPPA Proteomics
In addition, we perform primary analysis for other data types generated in Baylor core labs:
- Coding and noncoding transcriptomics: RNA-Seq, smallRNA-Seq
- Single cell transcriptomics scRNA-Seq
- Cistrome: ChIP-Seq, Reduced Representation Bisulfite Sequencing (RRBS), and Whole Genome Bisulfite Sequencing (WGBS)
- Genomics: Whole Genome Sequencing (WGS) or Whole Exome sequencing (WES)
Integrative Data Analysis
Integrative analysis typically involves pathway enrichment, computed using methods specific to each omics platform. Further, integration of different omics technologies, such as metabolomics and transcriptomics, or cistromics and transcriptomics, can be carried out. In addition to investigator data generated at the Baylor core facilities, we can assist with analysis and integration using publicly available datasets.
Data Deposition
Raw and processed data will be deposited in repositories such as GEO, Metabolomics Workbench. The core will collect from investigators sample metadata, fill in XML-format metadata sheets, and deposit the data and the metadata in public data repositories. Our goals are to meet or sometimes exceed state or federal mandated data deposition standards, to better serve the scientific community as well as enable rigorous data analysis reproducibility.
Facilities
The bioinformatics analysis will be performed using the infrastructure of the Dan L Duncan Cancer Center Computing Facilities:
In addition to up to date desktop computers for all faculty and staff, which include both 32-bit and 64-bit personal computers for most bioinformatically oriented members, we have two major computing facilities -- one in the Breast SPORE facilities on the main BCM campus and one on the Energy Transfer Data Center approximately one mile away. Both sit inside the BCM firewall, have nightly offsite backups, are protected through non-aqueous fire suppression, have redundant power and, for high and moderate capability machines, are accessible via a 10g Ethernet switched local area network (LAN).
The availability of two physically separated facilities dramatically improves availability by allowing for more rapid recovery from a disaster such as a fire or flood that incapacitates one facility. We have a 35 node high performance compute cluster (each with 2-8 cores each and newer nodes with 96 or 128 GB RAM) representing a total of 375 CPUs with 34 fast-access terabytes SAN storage for any high performance compute needs. We are in the process of expanding and substantially upgrading this capacity for 2013 with an extensible NetApp storage appliance.
For archival storage, 10s-100s of TB can be readily leased at very low cost from BCM Information Technology. Four cluster nodes are set aside for interactive jobs; the remaining nodes are available for batch jobs. Queues are managed by Sun Grid Engine and the system itself is administered by an expert system architect with >10 years of experience in HPC.
Access to these resources is supported by partial chargeback, commensurate with level of use. Because the cluster is self-contained (ie, located in one location without yet having an identical sister cluster offsite), the nodes themselves do not enjoy the full benefit of disaster recovery from both sites, whereas the archive storage does.
Outside of HPC availability, there are three Sun Sunfire X4170 Virtualization Servers at SUDC and two Cisco UCS C210 M2 Virtualization Servers at Breast facilities. Servers at both sites use VMware for creation of virtual servers that can run any operating system with varying system requirements. Each location’s virtualization servers have attached 37TB NetApp storage with vMotion in place to manage failover of the virtual machines from one site to another, should disaster situations arise.
In addition, there are two HP servers with direct-attached 96TB storage for Oracle 11g (backed up off-site nightly); four Sun physical servers with 37 terabytes of storage running the Solaris operating system; etc.
Charge Back Rates
Charge back rates for primary analysis of data generated by the CPRIT Core Facility are included as package in prices for each of the technology platforms. Charge back rates for secondary and integrative data analysis and other bioinformatics services are determined individually with investigators based on number of samples and analysis type.








