The Basics
The human microbiome, or microbiota, is defined as the collection of microbes - bacteria, viruses, and single-cell eukaryotes - that inhabits the human body. Microbes in a healthy human adult are estimated to outnumber human cells by a ratio of ten to one, and the total number of genes in the microbiome may exceed the number of genes in the human genome by a factor of about 100. Even though microbial cells are only one-tenth to one-hundredth the size of a human cell, they may account for up to five pounds of adult body weight.
Although bacteria are often associated with infections, the bacteria that colonize the surface and insides of our bodies are essential for life. We are dependent on these bacteria to help digest our food, produce certain vitamins, regulate our immune system, and keep us healthy by protecting us against disease-causing bacteria.
The identity and degree of diversity of the microbiota in a single individual is variable from person to person. The specific composition of a person's microbiome is increasingly recognized as playing an important role in disease and disease susceptibility.
The composition of the entire collection of microbes that make up the microbiome has, until recently, been difficult to investigate. Using traditional methods, it was not possible to isolate the vast majority (>80%) of microorganisms and culture them, presumably because the required growth conditions cannot or have not been able to be reproduced in the laboratory. However, technological advances in DNA sequencing and the development of a method known as metagenomics have made it feasible to analyze the entire human microbiome.
Metagenomics
Metagenomics is a sequence-based approach that allows the genetic material from the complete collection of microbes to be analyzed without needing to cultivate the microorganisms. Microbial communities can be harvested from their natural environments, and their DNA sequences can be determined, thereby providing a largely unbiased view of the human microbiome as well as the microbiomes of other animals and environmental niches.
The metagenomic approach allows for the identification of microorganisms that were previously unrecognized and gives vastly more information than the analysis of singly isolated microbes. Researchers can determine the relative abundance of the different species and discover which metabolic pathways are encoded by the organisms to gain information about their functions in the body. Metagenomic approaches have resulted in the identification of microbes and microbial functions that are associated with various stages of health and a variety of diseases.
The Microbiota in Health and Disease
The microbiome contributes to our health in diverse ways: by helping the body sense and respond to our environment, by harvesting nutrients from food, by playing a role in the development of our immune system, by preventing disease, and by controlling inflammation.
The microbiota may also influence many disease states, including diabetes, irritable bowel syndrome, and cancer, among others. Furthermore, the community of microbes that colonize the human body provides protection from incoming disease-causing microorganisms.
Through an understanding of how the microbiome impacts human health, researchers aim to translate this knowledge into new diagnostics and therapeutics.
The Research
In order to facilitate discovery of the roles that the microbiome plays in human health, nutrition, immunity, and disease, in 2007 the National Institutes of Health launched the Human Microbiome Project. Its mission was to generate the resources and expertise needed to characterize the human microbiome and analyze its role in health and disease. The NIH approved a budget of $170 million for this project over five years. This provided support for a number of centers and institutes around the United States, including one at Baylor College of Medicine, to serve as a "road map" for discovering the roles these microorganisms play in diverse niches of the human body.
The Alkek Center for Metagenomics and Microbiome Research (CMMR), based in the Department of Molecular Virology and Microbiology, was established in 2011 by Dr. Joseph F. Petrosino, a nationally recognized leader in metagenomic research, as an extension to Baylor's involvement in the Human Microbiome Project. The mission of the CMMR is to serve as an international hub for the development and implementation of advanced technologies for understanding how the microbiome impacts health and disease, and for the translation of this knowledge into microbiome-based therapeutics and diagnostics.
Researchers in the CMMR are developing molecular and bioinformatics tools and resources to advance numerous clinical and basic research projects pertaining to the organisms that comprise the microbiome, the genetic makeup of these bacteria, viruses and eukaryotes, and how this community of microorganisms interacts with human cells and tissues during the course of life.
The CMMR is actively involved in a great variety of projects that address the relationship between the microbiome and human disease, including irritable bowel syndrome, inflammatory bowel disease, Type 2 diabetes, leukemia, lung cancer, Crohn’s disease, among many others. A few of the research projects that focus on infectious diseases are summarized below.
The Microbiota and Resistance to Pathogenic Infections
In addition to the other functions of the microbiome in maintaining health, the community of microbes that make up the microbiota also provides resistance to colonization by dangerous, disease-causing microbes.
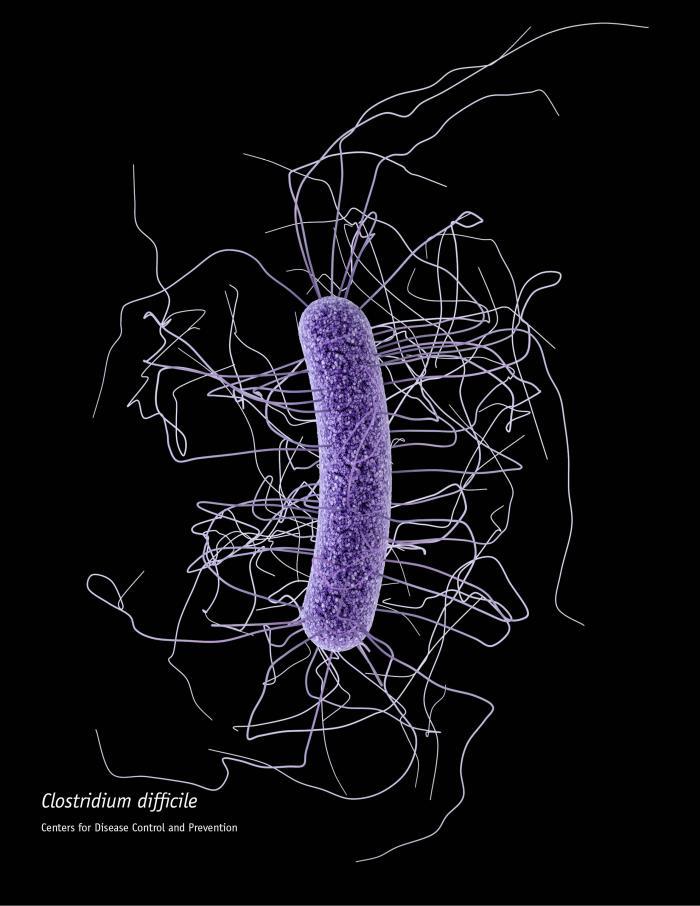
The laboratory of Dr. Robert Britton is interested in understanding how the intestinal microbiota provides a barrier to incoming pathogens and how perturbations of the microbiota result in an established infection. Most of the effort has been focused on the pathogen Clostridium difficile.
C. difficile is a gram-positive, spore-forming bacterium that is considered to be one of the three highest risk drug-resistant infections in the United States. It is an opportunistic infection that infects the colon of patients following antibiotic treatment. The microbiota that inhabit the gut normally prevent C. difficile colonization and suppress C. difficile-associated disease, but treatment with antibiotics results in changes to the composition of microbiota that allow C. difficile to grow and cause disease.
C. difficile produces toxins that damage intestinal cells and cause inflammation, producing diarrhea, and can be fatal. Of nearly 500,000 infections each year, approximately 29,000 result in death. It is the most common cause of hospital-acquired infections in developed countries. The infection is very difficult to treat, with many patients suffering from recurrent infections.
Dr. Britton, along with Dr. James Collins and other members of his research group, have developed mini-bioreactors and mice colonized with a human intestinal microbiota to determine which members of the microbial community are responsible for inhibiting C. difficile invasion. Their goal is to develop a probiotic cocktail, derived from the human intestinal microbiota, that will suppress C. difficile invasion.
Recently, C. difficile infections have become one of the most common causes of hospital acquired infections in the United States. This increase in infections was partially due to the emergence of epidemic, hypervirulent strains of C. difficile in the early 2000s, however the reasons for the emergence of these strains was unclear.
Using a combination of mini-bioreactors, a humanized mouse model, and bacterial physiology/genetics, the Britton laboratory was able to identify that two of the most common epidemic C. difficile ribotypes, RT027 and RT078, had acquired the ability to grow on the sugar trehalose at much lower concentrations that other C. difficile strains. They uncovered that these two ribotypes used two completely unique mechanisms by which they could now consume low quantities of trehalose.
Trehalose was approved for use in the human diet in 2000 by the FDA, immediately preceding the C. difficile epidemics that exploded in the early 2000s. The work of Dr. Britton and colleagues supports the idea that addition of this sugar to the human diet contributed to this outbreak.
Future work is aimed at understanding how other pathogens and the gut microbiota are impacted by recent changes to the human diet.
Impact of the Microbiota on the Immune System
Dr. Gretchen Diehl has been studying the impact of the microbiota on the immune system. Her research indicates that intestinal microbiota interact with the immune system to limit a damaging inflammatory response.
Infection by a disease-causing microbe, such as Salmonella, causes the body to mount a strong immune response that is directed at eliminating the microbe. The problem is that such an inflammatory immune response, especially in the intestine, can damage healthy tissue and contribute to conditions such as inflammatory bowel disease. Dr. Diehl and her coworkers have published a study that describes a cellular mechanism in which the microbiota play a role in modulating the immune response in a way that reduces inflammation and limits damage to the intestines.
The researchers found that in the presence of the normal microbiota, immune cells known as antigen-presenting cells regulated T-cell activity to balance the immune response. The microbiota instructed the antigen-presenting cells to secrete the cytokine IL-10. IL-10 served to dampen the response of inflammatory T helper cells that fight microbial invaders, while promoting the response of the anti-inflammatory regulatory T cells that are responsive to food antigens and the microbiota itself. The result is a balanced response that can fight off an infection, but prevents damage to healthy intestinal tissue.
When the laboratory animals used in their study were treated with antibiotics to eliminate the normal microbiota, the antigen-presenting cells did not produce IL-10. After re-populating the animals' intestines with bacteria, IL-10 production and a reduced inflammatory response were restored, but only if the bacteria could attach to the cells lining the intestines. Although microbes that attach to the intestinal epithelium are thought of as pathogens, the scientists found that attachment was necessary to produce a balanced immune response.
The results of this study suggest that manipulating the microbiota to limit intestinal immune responses could have potential therapeutic benefits for conditions such as inflammatory bowel disease. Dr. Diehl and her colleagues are searching for additional mechanisms by which microbes can balance immune responses and are investigating signaling pathways by which the microbiota can modulate immune responses.
Influence of the Microbiome on Neonatal Rotavirus Infection
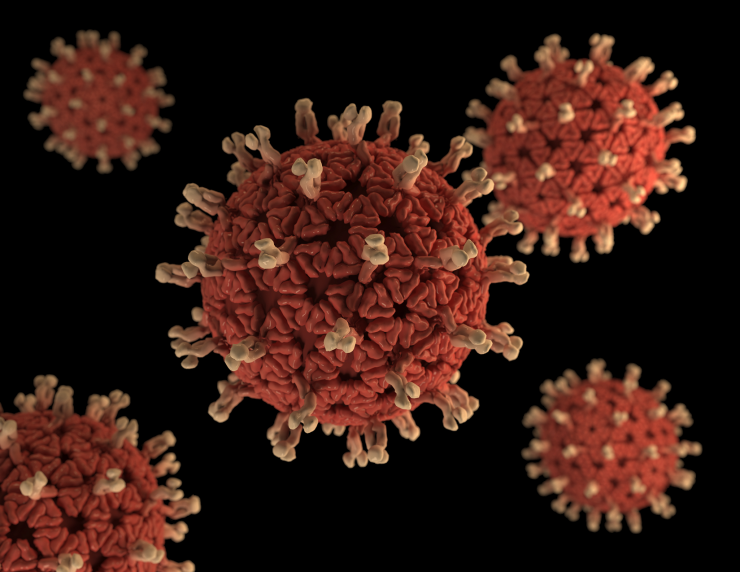
Rotavirus infection causes diarrhea and vomiting primarily in children younger than 5 years of age. While neonates (babies less than 28 days old) generally do not exhibit any symptoms, in some settings infections in newborns are associated with severe gastrointestinal disease. Researchers, including Dr. Sasirekha Ramani and Dr. Mary K. Estes, investigated factors that might influence whether newborns develop symptoms or remain asymptomatic.
The scientists studied a particular strain of rotavirus that they had identified in neonatal nurseries in India. This strain is associated with both severe gastrointestinal symptoms and infections without symptoms.
Previously, they tested whether factors such as the amount of virus in the newborns’ stool or the genome of the virus was linked to the presence or absence of symptoms, but they did not find any clear association.
They then investigated whether components in mother's milk could influence the outcome of the infection. Mother's milk contains many nutrients, including lipids, fats, proteins, and carbohydrates, small molecules that regulate immune development, as well as microbiota.
The results of their study indicated that a complex interaction between specific sugars in breast milk, the milk microbiome, and the infant gut microbiome impacts rotavirus infection in newborns. They found, unexpectedly, that with the particular neonatal strain of rotavirus, specific sugars in mother's milk enhanced infection in a cell line that is used as a model in rotavirus studies.
They further analyzed samples from mother-infant pairs and found that some of these same specific sugars are present in the milk of mothers of newborns with symptomatic infection, suggesting that specific sugars in breast milk can affect the outcomes of infections. This contrasted with previous work where sugars in breast milk reduced the infectivity of other rotavirus strains.
Since sugars in breast milk are important for the microbiome, the researchers characterized the breast milk and infant stool microbiomes from mother–infant pairs from both symptomatic and asymptomatic cases. Their analyses revealed that the relative abundance of certain types of bacteria (Enterobacter/Klebsiella) was significantly higher in the breast milk of mothers of symptomatic newborns compared to asymptomatic newborns or ones negative for rotavirus infection, while the levels of other types of bacteria (Staphylococcus and Streptococcus) were significantly lower. The scientists found similar results in their analysis of the infant fecal microbiome.
These data suggest that an interplay between sugars in mothers’ milk, the microbiome of breast milk, and the babies’ gut can influence symptomatic infections in neonates.
The role of breast milk components such as the milk microbiome in enteric infections is currently unexplored. These findings add to an increasing body of evidence that suggests a role of the microbiome in host-pathogen interactions and in influencing the outcome of infections.
Aberrant Microbe-host Communication Pathways Contribute to Rotavirus Disease
Interactions between microbes and their host use the same communication networks and pathways used to govern the day-to-day functions within our bodies. These communication pathways use an array of small molecules and cellular receptors to send and receive messages between cells and throughout the body. Some microbes co-opt these signaling systems to establish a stable home in our bodies, but many pathogens exploit these systems to benefit their growth and propagation. However, the aberrant nature of exploited signaling causes disease symptoms, including potentially life-threatening diseases like diarrhea.
Work in the laboratory of Dr. Joseph Hyser focuses on uncovering which of these communication pathways are important for supporting a healthy microbiome and which pathways are associated with diseases caused by pathogens, particularly viruses that cause diarrhea.
Most of the work uses time-lapse imaging of virus and bacterial infections of cells to watch and measure the change from normal to aberrant signaling using fluorescent sensors that are activated by important signaling molecules, such as Ca2+and cAMP. Using this technique, the scientists have discovered that rotavirus, a common diarrhea virus, activates hundreds of wide-spread aberrant signals through the secretion of ATP from a virus-infected cell to signal a neighboring uninfected cell.
Further, they have found that the extracellular ATP signal significantly contributes to the generation of rotavirus diarrhea. Therefore, new therapies that block this communication pathway may reduce rotavirus disease burden or deaths.
Extracellular ATP signaling is an ancient form of cellular communication, and rotavirus is the first virus identified to exploit this signaling pathway to increase its replication and cause disease. However, the Hyser Lab expects to identify other diarrhea-causing pathogens that exploit this or related pathways.
Glossary
Learn more about some of the technical terms found on our Emerging Infections and Biodefense pages.








 Credit
Credit